How to find the nucleotide sequence for your gene (ATCG stuff)
A beginners find to finding the genomic DNA sequence of your gene.
TL;DR
I wrote a simple guide for how to find the sequence for a gene
I also built a web app that simplifies the process
https://searchgene.netlify.app/
I have restricted the web app to only human genes for now
Assumptions for this post
You are in the early stages of learning biology
You are trying to teach yourself biology
You aren’t familiar with biology jargon
You don’t have a lot of experience reading and implementing biology papers
You need clear, written instructions on how to do things (even “basic” things)
You know the gene you are searching for, including the name and the origin (human, mouse etc)
Important things of note about this post
I am not writing in a lot of detail. Nor am I writing precisely or for engagement purposes.
The aim extreme is extreme clarity for absolute beginners so that they can do this stuff on their own.
Hence only short sentences and bullet point notation is guides like these..
What is a nucleotide sequence?
It is the sequence made up of A (Adenine), T (Thymine), C (Cytosine), and G (Guanine)
There are different types of nucleotide sequences (genomic DNA and mRNA being the two you will use the most)
Genomic DNA is used to create mRNA.
mRNA is used to create proteins.
You can think of A being opposite to T, and C being opposite to G
The genomic DNA sequence is the sequence actually found in your DNA. This is what provides the template for your mRNA to be made.
The mRNA sequence is opposite to the genomic DNA sequence.
So if the genomic DNA sequence has AATC, the mRNA sequence will have TTAG.
This is all you need to know for now to find the genomic DNA sequence of your gene
How to find your genomic DNA sequence
Find the name of your gene (ideally the shorthand version of the name, also known as the gene symbol e.g Sox2 is the shorthand version aka Gene Symbol for SRY (sex determining region Y)-box 2
Go to https://www.ncbi.nlm.nih.gov/
Choose the gene database (where it initially says “All databases” select the dropdown and change it to “Gene”)
Type in the name of the gene of interest (I am using SOX2 as an example)
Scroll down the search results.
Check the description box for its source of origin (Homo sapiens = Humans, Mus Musculus = Mouse. This should be stated in brackets next to the gene name ).
Look in the description column and select the gene you are looking for. In our example, we would choose the second result as we are looking for the human gene. (“[ Homo sapiens (human) ]”)
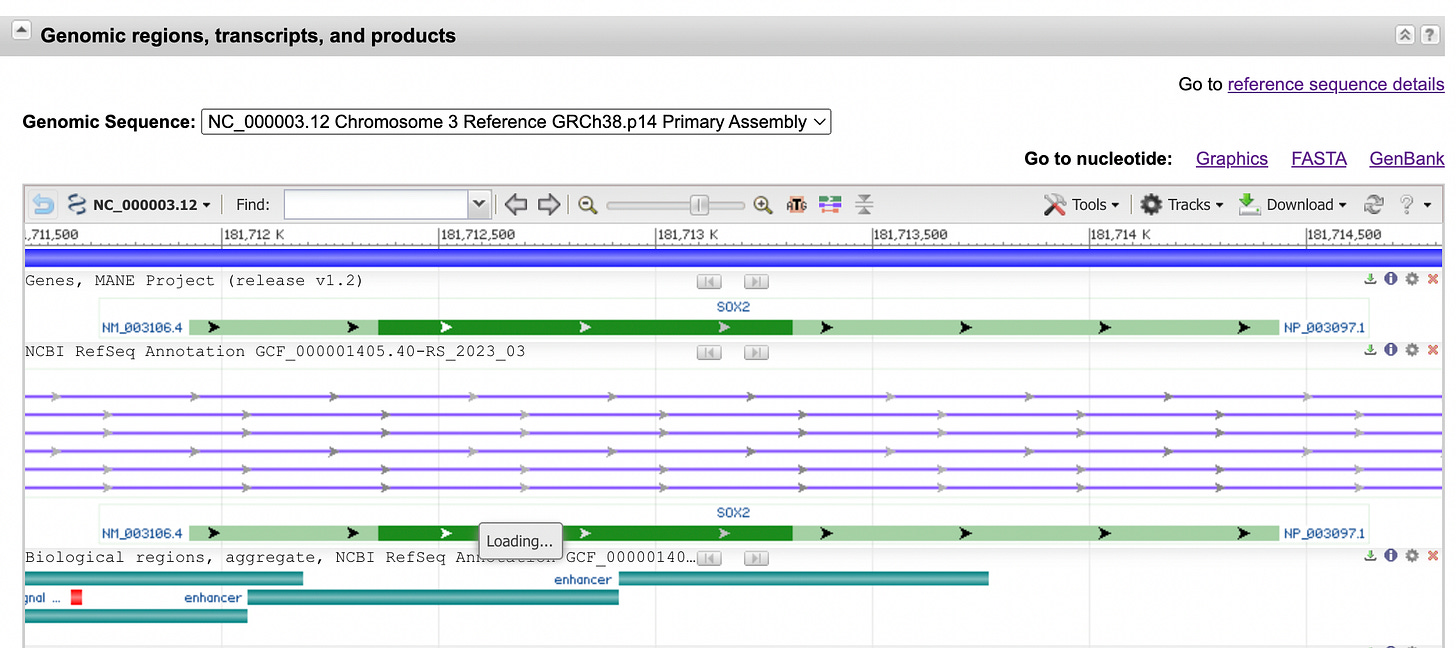
Scroll down to “Genomic regions, transcripts, and products” section
Select FASTA (where it says “Go to nucleotide”, there are 3 options next to it).
You have your sequence!
What things mean
The “Genomic Sequence” dropdown. Using our example of Sox2, when you are in the “Genomic regions, transcripts, and products” section you will see a dropdown bar that is called “Genomic Sequence” with a strange-looking number similar to this “NC_000003.12 chromosome 3 Reference GRCh38.p14 Primary Assembly”. This is what each section means (courtesy of ChatGPT):
**NC_000003.12**: This is a unique identifier for a specific version or release of the DNA sequence of human chromosome 3. It ensures that scientists are referring to the exact same "edition" or version of the chromosome's DNA sequence when discussing or researching it.
**chromosome 3**: This indicates that the DNA sequence in question belongs to the third of the 23 pairs of chromosomes in humans. Each chromosome contains a large number of genes, and this specifies which one we're discussing.
**GRCh38.p14**: This is the specific version of the entire human genome reference sequence. "GRCh38" tells us it's the 38th version coordinated by the Genome Reference Consortium.
The "p14" indicates that this is the 14th patch or minor update to that version. Think of this as the equivalent to minor bug fixes on an app e.g v.0.0.1, v.0.0.1.2 etc
**Primary Assembly**: This means that the sequence is the main or default representation of the human genome. It serves as the standard to which other genomic data can be compared or aligned. The primary assembly provides a continuous, standard sequence for each chromosome or genomic region, even if there might be variations or alternate versions in reality.
So, putting it all together, This is a specific version of the DNA sequence for human chromosome 3, which is part of the main or standard version of the 38th edition of the human genome reference.
Extra things you may see:
**NC_000003.12 (181711925..181714436)**: This is the location of the sequence. In our case it means means human chromosome 3 in a particular version of its sequence.
"(181711925..181714436)": These numbers define a specific range or segment on chromosome 3. This means you're looking at a section of the DNA that starts at position 181,711,925 and ends at position 181,714,436. The segment in question is, therefore, 2,511 bases (or "letters") long.
Bonus
I created a simple web app to search for human genes without doing all of the steps mentioned above.
Here is the link https://searchgene.netlify.app/
It always chooses the latest version of a gene (although that may not be what you want!).
I have also limited it to only finding genomic DNA sequences (no mRNA)
It only finds human genes (worth mentioning again).
I plan on adding more features later, but this is purely a side thing to help beginners out.
If I got anything wrong, or you need any help with anything, lmk!